XU LABORATORY
INSTITUTE FOR CELL ENGINEERING

DEPARTMENT OF NEUROLOGY | JOHNS HOPKINS SCHOOL OF MEDICINE


Brain resilience
&
Energy control
Transcription and brain resilience
Unlocking the Brain’s Hidden Resilience Switch:
— For decades, Alzheimer’s therapeutics have emphasized clearing amyloid plaques and tau tangles after they accumulate, yet biomarker improvements have seldom translated into meaningful cognitive recovery or durable slowing of decline. This disconnect argues that AD is not merely an aggregate-removal problem, but a progressive breakdown of neural communication: synapses fail, network rhythms destabilize, and excitation–inhibition balance erodes. Our strategy shifts from reacting to late-stage pathology to proactively restoring intrinsic circuit resilience, synaptic organizers that stabilize specific neuronal interactions and supports efficient information flow. To interrogate this mechanism in a human context, we engineered iPSC-derived, E/I-balanced cortical networks (including rhythm-critical inhibitory interneurons) and immune-vascularized forebrain organoids, enabling direct functional readouts of synaptic strength and oscillatory dynamics. We find synaptic organizer loss reflects regulatory shutdown—impaired gene activation and protein production—rather than gene deletion, motivating precision approaches to reopen this resilience program.
Our research has revealed that following the initiation of pro-resilience activity in human iPSC derived brain cells, PRDM gene family members, play crucial roles in the transitions of cellular states between brain cell resilience and demise. This discovery significantly broadens our understanding of the PRDM gene family. These transcription factors, known for either acting as direct histone methyltransferases or recruiting a range of histone-modifying enzymes, target specific promoters in defined contexts. This targeting is pivotal for driving and maintaining the delicate equilibrium in cellular state transitions and for modulating the activity of signaling pathways.
Specifically, our lab is exploring the role of PRDM gene family in AD, focusing on their regulation of:
— Vascular endothelial permeability and integrity
— Astrocyte metabolism
— Cellular crosstalk driven by the PRDM genes
— Neutrophil stall in capillaries
Translation is dynamic
Over the course of development, over the course of an experience, or over the course of disease development, how a brain cell translates its transcriptome charts its fate and function. The translation of specific mRNA can be dynamically regulated by signaling pathways, nutrient availability, misfolded protein accumulation, and brain cell activity.
In the lab, we have deployed modern molecular techniques to understand how translation control mechanisms operate in neural cells and how their disruption causes neurological disorders.
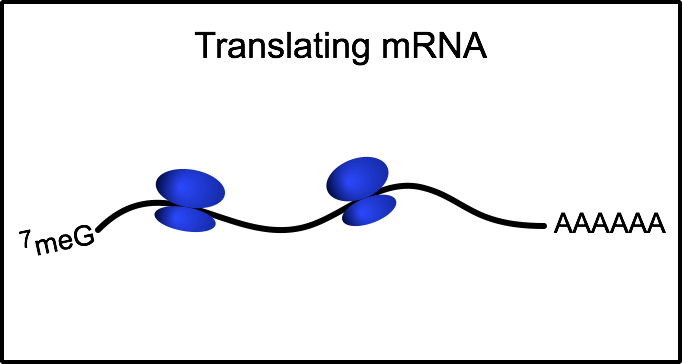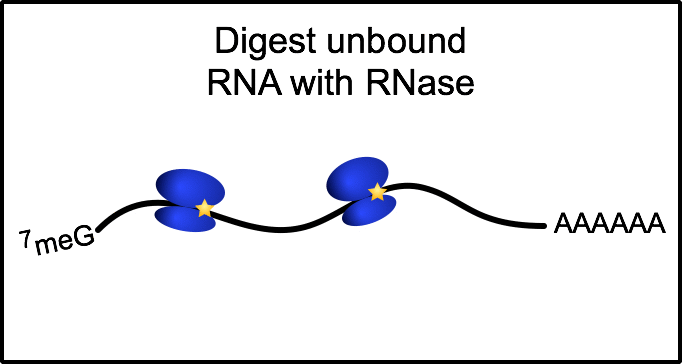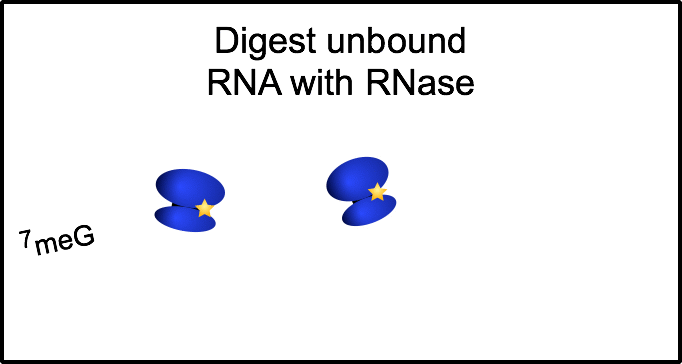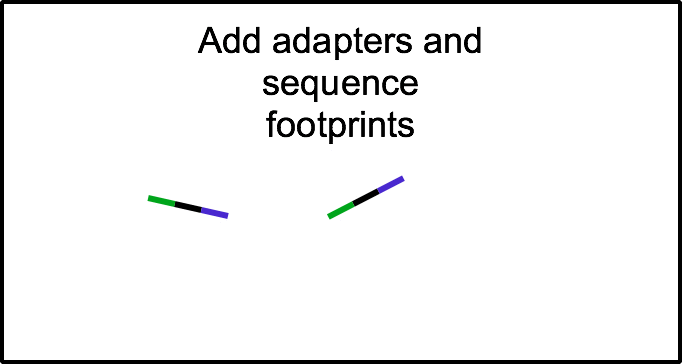
Ribosome profiling: a quantitative method to assess translation
To gain a genome-wide perspective on how neural cells regulate translation, we employ a method called ribosome profiling. Following a mild RNase digestion, we recover and sequence ribosome protected fragments (or footprints). Comparing with total mRNAseq from the same sample, we can derive the efficiency at which any given mRNA is translated.
mTOR, human neural precursor & Alzheimer disease
We have applied ribosome profiling to human embryonic stem cell derived neural precursor cells (hNPCs) to understand how the mechanistic target of rapamycin (mTOR) regulates the specification of subtypes of precursors.
Dysregulation of mTOR signaling underlies numerous neurological disorders including Alzheimer, autism etc. Our results so far point to an important role for mTOR-mediated translation control in the specification a particular subtype of hNPCs and Alzheimer related genes by targeting unconventional genes with either TOP-like sequences or no discernible TOP sequence.


I.Wu
Cell-type specific Ribosome profiling
in vivo
Understanding how translation is regulated within the intact nervous system presents enormous challenges, not least of which is the diversity of cell types within any given structure in the brain.
Combining the translating ribosome affinity purification method with ribosome profiling (described above) we have developed method that allows us to study translation in genetically defined cell types in the mouse brain. This method allows us to determine how translation is altered by behaviorally relevant stimuli in specific cell types. In combination with targeted mutations , we can dissect how translation control machinery regulates what proteins are synthesized in neurons.
Ribosome profiling gives us a nucleotide level resolution of translation. This resolution allows us to identify translated segments of the genome not previously characterized including non-canonical open reading frames upstream and downstream (uORFs and dORFs) of the canonical main open reading frame (mORF).

S, Eacker
